SAM-TB
Home
My data
My analysis
My samples
Help
FAQ
中文
Sign in
##Core genome multilocus sequence typing (cgMLST) Core Genome Multilocus Sequence Typing (cgMLST) is an efficient, easily standardized, and storage-saving genotyping method with the potential to monitor large-scale tuberculosis transmission. Our website uses the MTBC cgMLST scheme designed by Kohl T.A. et al[1] for genotyping. This scheme uses 2891 core genes to determine the cgMLST genotype of each strain and standardizes strain typing through the online [cgMLST allele nomenclature system](http://www.cgmlst.org/mtbc). ###Operating instructions The cgMLST typing function has been integrated into the “Single Sample Variants Analysis” module. When users initiate this module for analysis, the website will automatically convert the identified whole-genome variants into a cgMLST allele profile. For further information on performing a “Single Sample Variant Analysis”, users can refer to the **[Single Sample Variant Analysis](/docs/singlesample)** section in the Help Center. ###Explanation of results (1) After the analysis is completed normally, click on the analysis number to view the results of the sample. The “cgMLST” page in the secondary navigation bar will display the cgMLST allele profile of the sample. As shown in the figure below, the first column displays the name of the cgMLST gene; the second column lists all the variations on the corresponding gene; and the third column shows the genotype of that gene, represented by the standardized numerical codes provided at [cgMLST allele nomenclature system](http://www.cgmlst.org/mtbc). 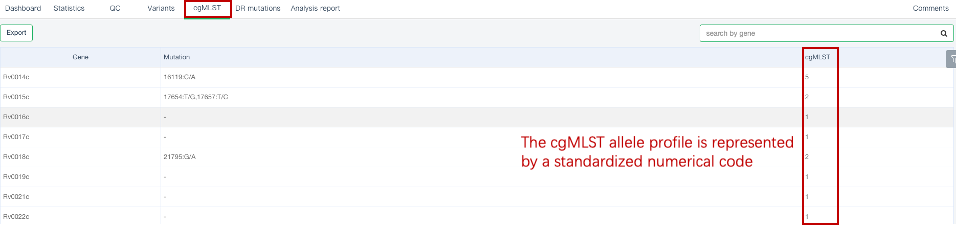 (2) Our website supports the batch export of the integrated cgMLST allele profile matrix for samples that have completed analysis. In this matrix, rows represent samples, columns represent genes, and the values in the cells indicate the genotypes. For specific operation methods, please refer to the **[Quick Analysis](/docs/multisample)** section in the Help Center, where detailed step-by-step instructions are provided in the **View and download analysis results** subsection. ###Reference [1] [Kohl TA, Harmsen D, Rothgänger J, Walker T, Diel R, Niemann S (2018) Harmonized genome wide typing of tubercle bacilli using a web-based gene-by-gene nomenclature system. EBioMedicine 34:131–138.](https://doi.org/10.1016/j.ebiom.2018.07.030)
<< Return
Title:
Description:
Thank you for using our service, we will reply you by email as soon as possible.