SAM-TB
Home
My data
My analysis
My samples
Help
FAQ
中文
Sign in
##Identification of mixed MTB infections When a host is infected with two or more strains of Mycobacterium tuberculosis (MTB) at the same time, an MTB mixed infection occurs. Accurately determining the presence of MTB mixed infection is crucial for assessing treatment outcomes and reconstructing transmission among patients. Our website integrates the Mixinfect tool developed by Sobkowiak, B., et al. in 2018[1]—using Bayesian model clustering techniques to detect whether a sample contains a mixed infection of two or more MTB strains. ###Operating instructions The identification function for MTB mixed infection has been integrated into the “Single Sample Variants Analysis” module. When users initiate this module for analysis, the website will automatically perform mixed infection detection on all samples identified as MTB, while samples identified as NTM will not be included in this detection. For further information on performing a “Single Sample Variant Analysis”, users can refer to the **[Single Sample Variant Analysis](/docs/singlesample)** section in the Help Center. ###Explanation of results (1) After the analysis is completed normally, click on the analysis number to view the results of the sample. In the “Dashboard” section of the secondary navigation bar, the “Analysis Summary” will display the mixed infection identification of the sample. For example, the results shown in the figure below indicate that the sample has a mixed infection of two MTB strains, with the estimated proportion of the main strain being 81.47%. If only one MTB strain is detected in the sample, the result will be marked as “Not mixed infection”. 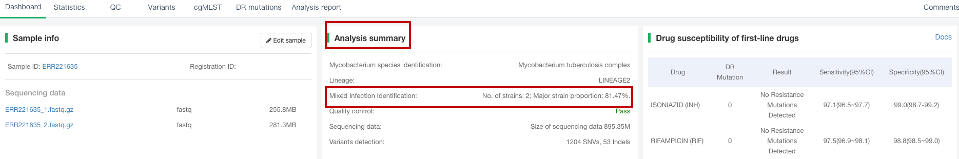 (2) Our website also offers a batch export function, allowing users to export the results of mixed infection identification for samples that have completed analysis. For specific operation methods, please refer to the **[Quick Analysis](/docs/multisample)** section in the Help Center, where detailed step-by-step instructions are provided in the **View and download analysis results** subsection. ###Reference [1] [Sobkowiak, B., Glynn, J.R., Houben, R.M.G.J. et al. Identifying mixed Mycobacterium tuberculosis infections from whole genome sequence data. BMC Genomics 19, 613 (2018).](https://doi.org/10.1186/s12864-018-4988-z)
<< Return
Title:
Description:
Thank you for using our service, we will reply you by email as soon as possible.